Niepielko Lab
Dr. Matthew G. Niepielko is an Assistant Professor of Computational Biology and Activities Coordinator for the Group Summer Scholars Research Program for high school students at Kean. His laboratory investigates the molecular mechanisms that regulate animal development and fertility using different species of Drosophila (fruit flies) as a model system.
Matthew G. Niepielko's CV Members
About
Dr. Matthew G. Niepielko is an Assistant Professor of Computational Biology and Activities Coordinator for the Group Summer Scholars Research Program for high school students at Kean. His laboratory investigates the molecular mechanisms that regulate animal development and fertility using different species of Drosophila (fruit flies) as a model system. Dr. Niepielko has identified several developmental mechanisms that mediate the evolution of morphologies during Drosophila eggshell formation. He has also discovered key mechanisms that regulate cell polarity during Drosophila development by employing a novel technique that couples biological experiments with computational analyses. The Niepielko Lab currently explores how germ cell mRNA composition alters animal fertility and how changes in gene expression drives evolution.
Dr. Niepielko received his Ph.D. in Computational and Integrative Biology from Rutgers University in 2014. In 2016, he received the Ruth L. Kirschstein National Research Service from the National Institutes of Health to complete his postdoctoral training at Princeton University. In 2021, he was awarded an Academic Research Enhancement Award (AREA) from the National Institutes of Health to support research activities at Kean University. In 2023, Dr. Niepielko was awarded the Faculty Early Career Development Program Award (CAREER Award) that will fund his research and STEM education program, New Jersey's Research Alliance for Inclusive STEM Education (NJ-RAISE), which focuses on including high school students and undergraduates in STEM research training and education opportunities.
Announcements
4/22/2025
Congratulations to Niepielko Lab alumna Katie Morrison on her acceptance and commitment to the Philadelphia College of Osteopathic Medicine! Looking forward to your success!
3/20/2025
Congratulations to Kristina Spencer for being accepted to an REU program this summer at Boston University! What an amazing opportunity!
3/12/2025
Congrats to Ahad Shabazz-Henry for receiving the Undergraduate Researcher of the Year award in Natural Sciences! What a HUGE accomplishment!
Previous Announcements: Niepielko Lab News
Research
Investigating mechanisms that regulate reproductive fitness in Drosophila
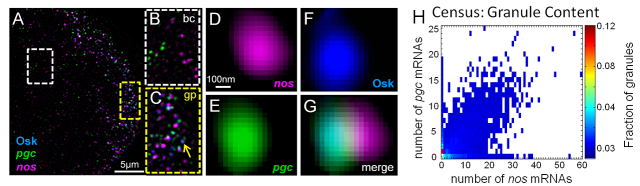
A fundamental requirement for animal reproduction is the development and maintenance of the germline, the set of highly specialized cells responsible for passing on genetic material to the following generation. Recent discoveries have revealed that germline function requires the formation of highly conserved ribonucleoprotein (RNP) granules called germ granules. Germ granules have essential roles in germline differentiation, proliferation, and post-transcriptional gene regulation. In animals such as Drosophila, germ cell specification occurs through the inheritance of germ granules that reside at a specific location within the egg. In humans, where germline fate is induced through signaling interactions, similar germ granules form de novo after germline differentiation. Regardless of their origin, germ granules contain highly conserved components such as mRNA encoding the translational repressor nanos (nos). Evidence supporting the conserved role of germ granules comes from the effect of mutations that eliminate conserved germ granule components in Drosophila, Xenopus, zebrafish, and mouse, which result in the loss of the germline. In humans, mutations in the nos ortholog, NANOS1, are associated with defects in spermatogenesis that result in a lack of germ cells in the testes, while mutations in NANOS3 are linked to premature ovarian failure. Despite the conserved function of germ granules, it is unclear how fertility may be affected by changes in germ granule composition. Elucidating how granule mRNA content affects fertility and the mechanisms that yield germ granule diversity should provide insight into defects such as infertility and sterility. In the Niepielko Lab, we take advantage of the natural variation of fertility found in Drosophila species to investigate the role germ granules have in reproductive robustness.
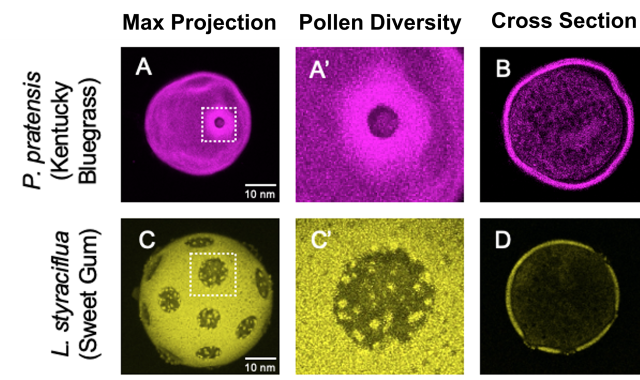
Capturing 3D pollen images using confocal microscopy
Seasonal allergic rhinitis (SAR) is a common inflammatory condition caused by pollen grains released by trees, grasses, weeds, or molds. Many people are affected by the cold-like symptoms caused by these various pollen species. Therefore, streamlining the distribution of real-time pollen conditions is important because it can provide allergy sufferers with useful information to help reduce pollen exposure. AI can be used to automate the process of identifying and quantifying real time pollen conditions. It is hypothesized that confocal microscopy images can serve as sufficient training data for this AI program. Whereas other types of microscopes can only allow the external characteristics of samples to be seen, confocal microscopy allows a sample to be imaged in slices along its z-axis which are then used to create 3D and cross-sectional images. This allows the images to not only display the external morphologies and characteristics that are unique to each pollen species, but also the internal morphologies and characteristics. During this study, several 3D images of various pollen species were captured at various magnifications. The images were able to show and distinguish each pollen species by their distinctive characteristics. The study found that confocal microscopy can be used to produce detailed images of pollen grain species. The next step in this research would be to develop an AI program that can identify and quantify pollen on an unmodified slide of current pollen conditions. This information can then be made public online or added to a pollen database.